DNA details to cash-in on CRISPR-Cas
 A new paper has described some details of a genome-editing system that is quickly becoming the global standard.
A new paper has described some details of a genome-editing system that is quickly becoming the global standard.
Customised genes built by editing DNA sequences to add, delete, activate or suppress specific sequences have major potential applications in almost all fields.
In a new paper published in Molecular Cell, researchers examine six key molecular elements that help drive CRISPR-Cas - a hugely popular genome editing system.
CRISPR stands for “clustered regularly interspaced short palindromic repeats,” and Cas is a family of genes and corresponding proteins associated with the CRISPR system, which specifically target and cut DNA in a sequence-dependent manner.
Essentially, the authors say, bacteria use the system as a defence mechanism and immune system against unwanted invaders such as viruses.
That same system is now being harnessed by researchers to quickly and more precisely target certain genes for editing.
“This paper sheds light on how CRISPR-Cas works,” North Carolina State University researcher Dr Rodolphe Barrangou said.
“If we liken this system to a puzzle, this paper shows what some of the system’s pieces are and how they interlock with one another. More importantly, we find which pieces are important structurally or functionally – and which ones are not.”
Barrangou says that the paper’s findings will allow researchers to increase the specificity and efficiency in targeting DNA, setting the stage for more precise genetic modifications.
An abstract of the paper is presented below:
“Guide RNA Functional Modules Direct Cas9 Activity and Orthogonality”
Authors: Alexandra E. Briner, Kurt Selle, Chase L. Beisel and Rodolphe Barrangou, North Carolina State University; Paul D. Donohoue, Euan M. Slorach, Christopher H. Nye, Rachel E. Haurwitz, and Andrew P. May, Caribou Biosciences Inc.; Ahmed A. Gomaa, University of Cairo
Published: Oct. 16, 2014, online in Molecular Cell
DOI: 10.1016/j.molcel.2014.09.019
Abstract:
The RNA-guided Cas9 endonuclease specifically targets and cleaves DNA in a sequence-dependent manner and has been widely used for programmable genome editing.
Cas9 activity is dependent on interactions with guide RNAs, and evolutionarily divergent Cas9 nucleases have been shown to work orthogonally. However, the molecular basis of selective Cas9:guide-RNA interactions is poorly understood.
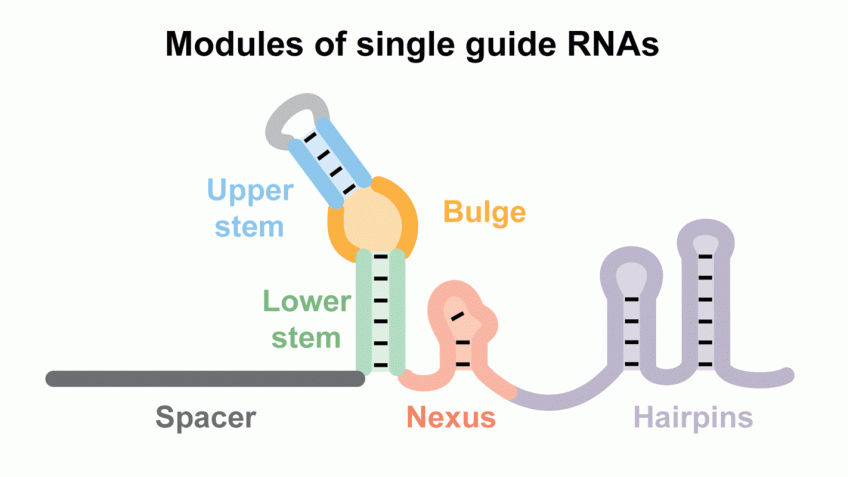
Here, we identify and characterize six conserved modules within native crRNA:tracrRNA duplexes and single guide RNAs (sgRNAs) that direct Cas9 endonuclease activity.
We show the bulge and nexus are necessary for DNA cleavage and demonstrate that the nexus and hairpins are instrumental in defining orthogonality between systems.
In contrast, the crRNA:tracrRNA complementary region can be modified or partially removed.
Collectively, our results establish guide RNA features that drive DNA targeting by Cas9 and open new design and engineering avenues for CRISPR technologies.







 Print
Print